dataset - Is there a way to convert smiles format to TUdataset
$ 29.50 · 4.9 (291) · In stock


PDF) Design of a Chemical Graph Database to Query on an Atom-Specific Level

PDF) EPIC: Graph Augmentation with Edit Path Interpolation via Learnable Cost

PDF] Multi-caption Text-to-Face Synthesis: Dataset and Algorithm
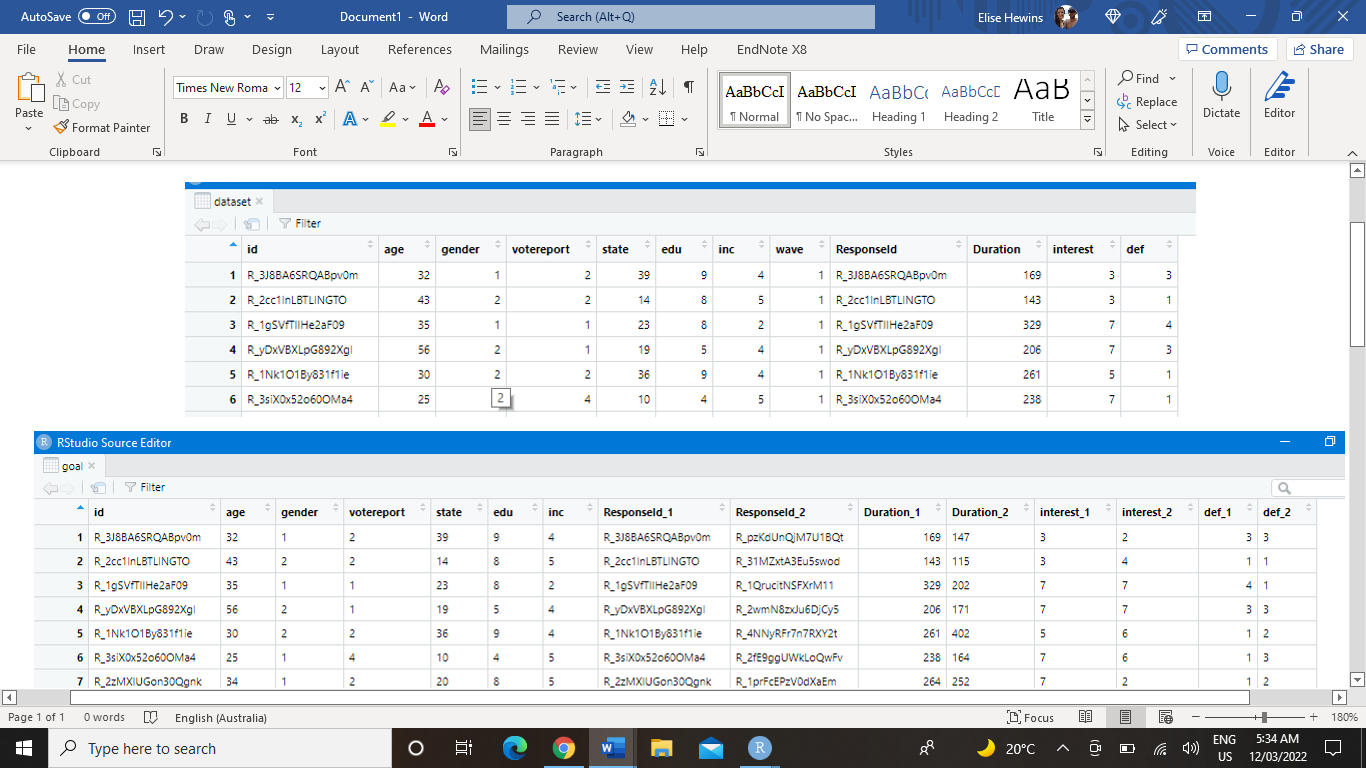
converting a dataset from a long to wide format. - tidyverse - Posit Community

2103.09430] OGB-LSC: A Large-Scale Challenge for Machine Learning on Graphs

Relating the xyz files to the molecules in PCQM4Mv2 · Issue #297 · snap-stanford/ogb · GitHub

PDF) Hierarchical Grammar-Induced Geometry for Data-Efficient Molecular Property Prediction
Pytorch-Geometric/pytorch_geometric_introduction.py at master · marcin-laskowski/Pytorch-Geometric · GitHub
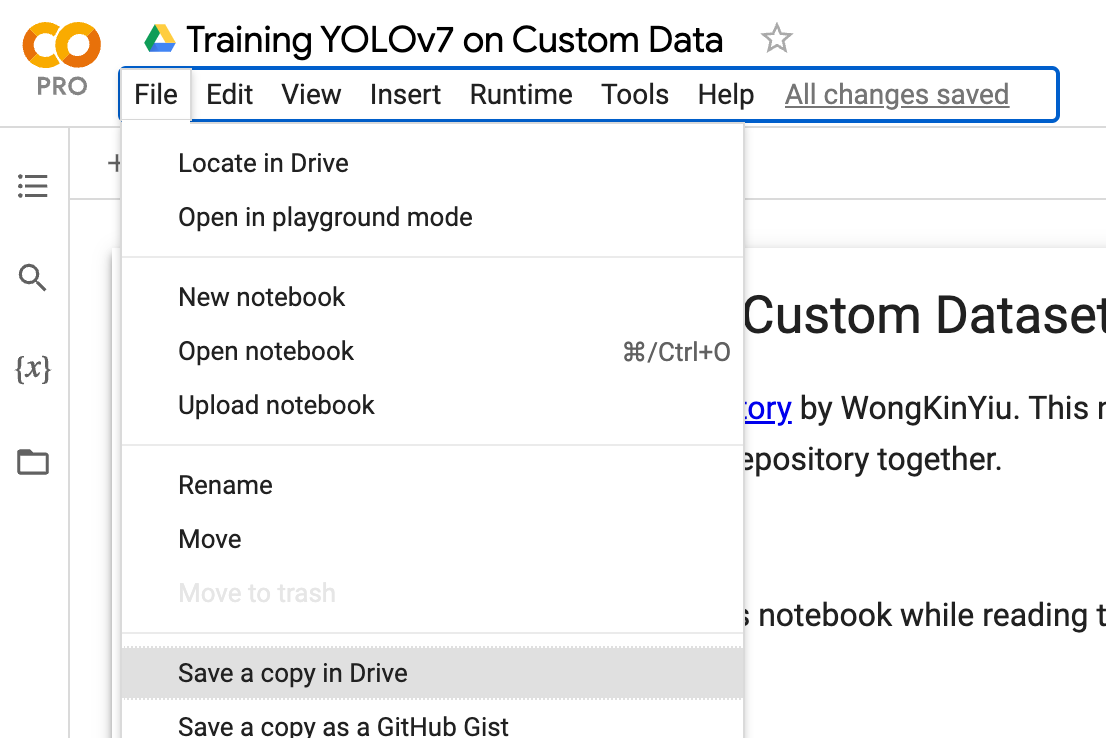
How to Train YOLOv7 on a Custom Dataset

PDF) Visual Analytics For Machine Learning: A Data Perspective Survey

Metapath-fused heterogeneous graph network for molecular property prediction - ScienceDirect
sdftosmi.py: Convert multiple ligands/compounds in SDF format to SMILES. — Bioinformatics Review
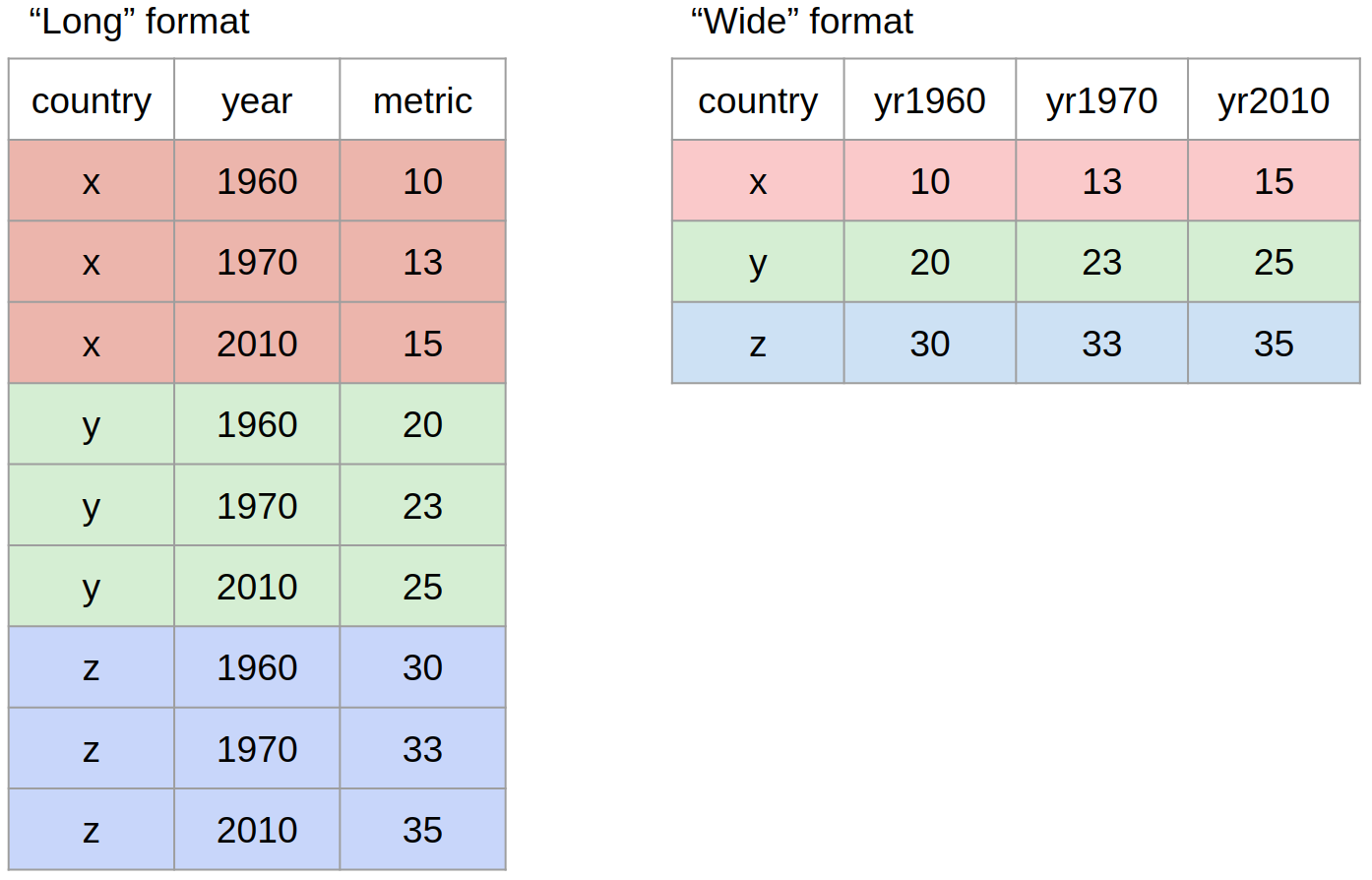
Data reshaping: from wide to long and back – Introduction to R/tidyverse for Exploratory Data Analysis